pSCoPE
Prioritized single-cell proteomics method by Huffman et al, 2022
- Code available at: github.com/SlavovLab/pSCoPE
- Peer reviewed article: Huffman RG, Leduc A, Wichmann C, … and Slavov N, Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics. Nat Methods, doi: 10.1038/s41592-023-01830-1 (2023)
- Research Briefing: Extending the sensitivity, consistency and depth of single-cell proteomics, OA
Data Websites
- Huffman et al., 2022
- Leduc et al., 2022
- Leduc et al., 2023
- Leduc et al., 2024
- Khan, Elcheikhali et al, 2024
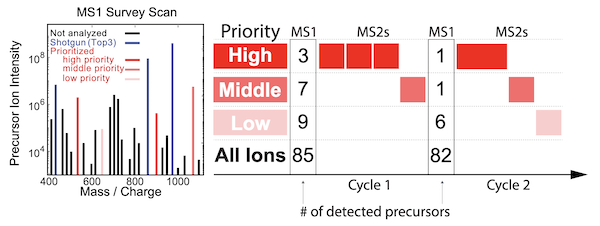
Major aims of single-cell proteomics include increasing the consistency, sensitivity, and depth of protein quantification, especially for proteins and modifications of biological interest. To simultaneously advance all of these aims, we developed prioritized Single Cell ProtEomics (pSCoPE). pSCoPE ensures duty-cycle time for analyzing prioritized peptides across all single cells (thus increasing data consistency) while analyzing identifiable peptides at full duty-cycle, thus increasing proteome depth. These strategies increased the quantified data points for challenging peptides and the overall proteome coverage about 2-fold.
Implementation
pSCoPE is implemented as a module of MaxQuant.Live, starting with version 2.1 and is compatible with Thermo Fisher Q-Exactive series, Orbitrap Exploris as well as Orbitrap Eclipse. All additional code needed for pSCoPE and reproducing Huffman et al., 2022 is available at github.com/SlavovLab/pSCoPE. Try it out and give us feedback!