QuantQC
Quantitative evaluation and processing pipeline by Leduc et al, 2023
- Code available at: github.com/SlavovLab/QuantQC
- Preprint Protocol: Leduc A, Luke Khoury, Joshua Cantlon, Slavov N. (2023) Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP, bioRxiv, doi: 10.1101/2023.11.27.568927
- Peer-reviewed Protocol: Leduc A, Luke Khoury, Joshua Cantlon, Slavov N. (2024) Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP, Nature Protocols, doi: 10.1038/s41596-024-01033-8, blog
- Peer-reviewed Article: Leduc et al., Limiting the impact of protein leakage in single-cell proteomics, Nature Communications doi: 10.1038/s41467-025-56736-7
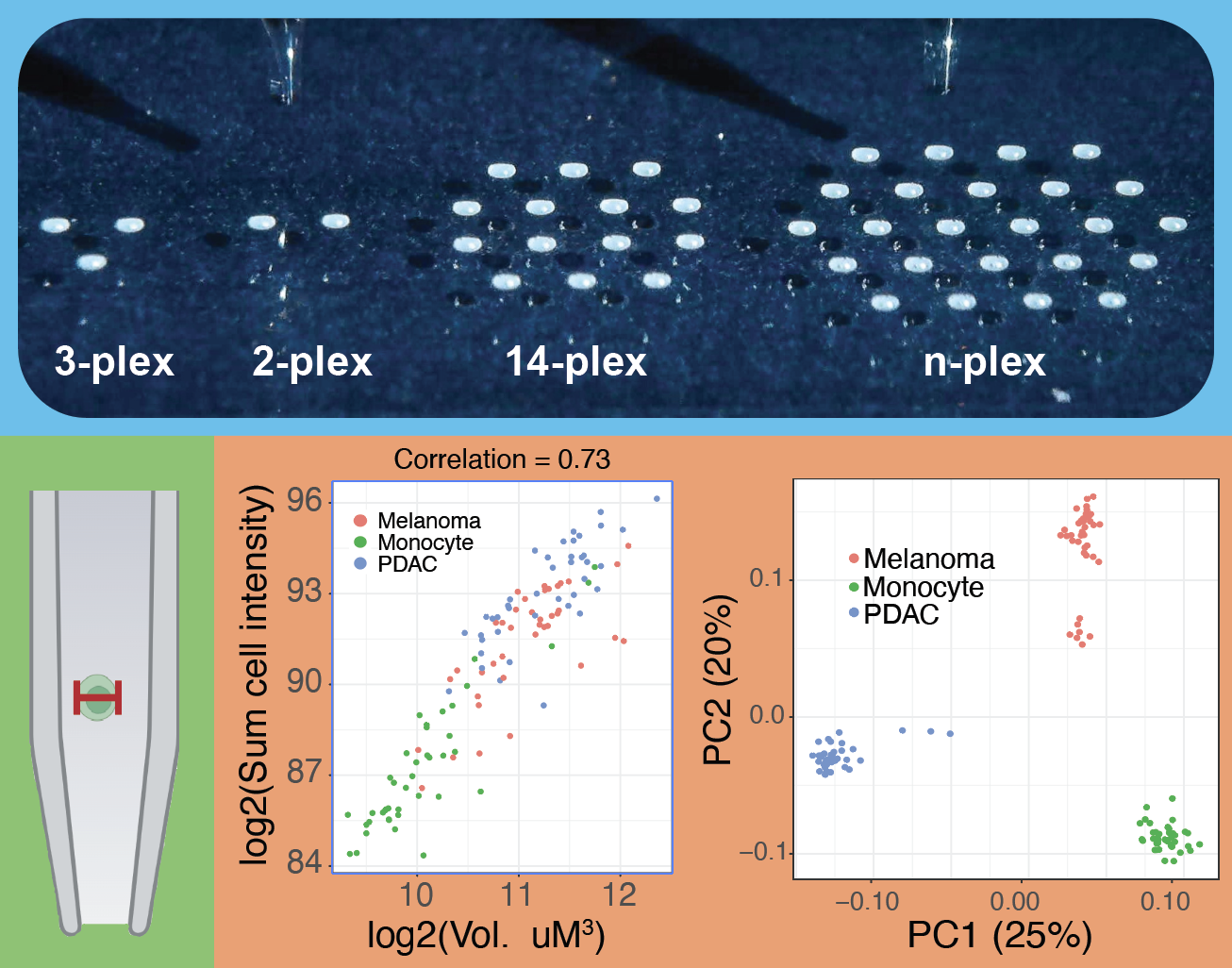
QuantQC is an R package for evaluation, benchmarking and processing of single-cell proteomics data, especially data from experiments using nPOP. It can be used to map all metadata and generate quality reports for quick evaluation of the experiment. QuantQC generates HTML reports for evaluating nPOP sample preparation, stability of data acquisition, and quantification performance that can be easily shared with colleagues. QuantQC also facilitates exploratory data analysis such as visualizing agreement between peptides mapping to the same protein across clusters.
Implementation
QuantQC is available as an R package from github.com/SlavovLab/QuantQC. Try it out and give us feedback!