Minimal Proteomic sample preparation (mPOP)
Sample prep method by Specht et al, 2018
mPOP Article Nature Protocol JoVE Protocol
Data Websites
Introduction Advantages of mPOP RAW Data Videos
Introduction
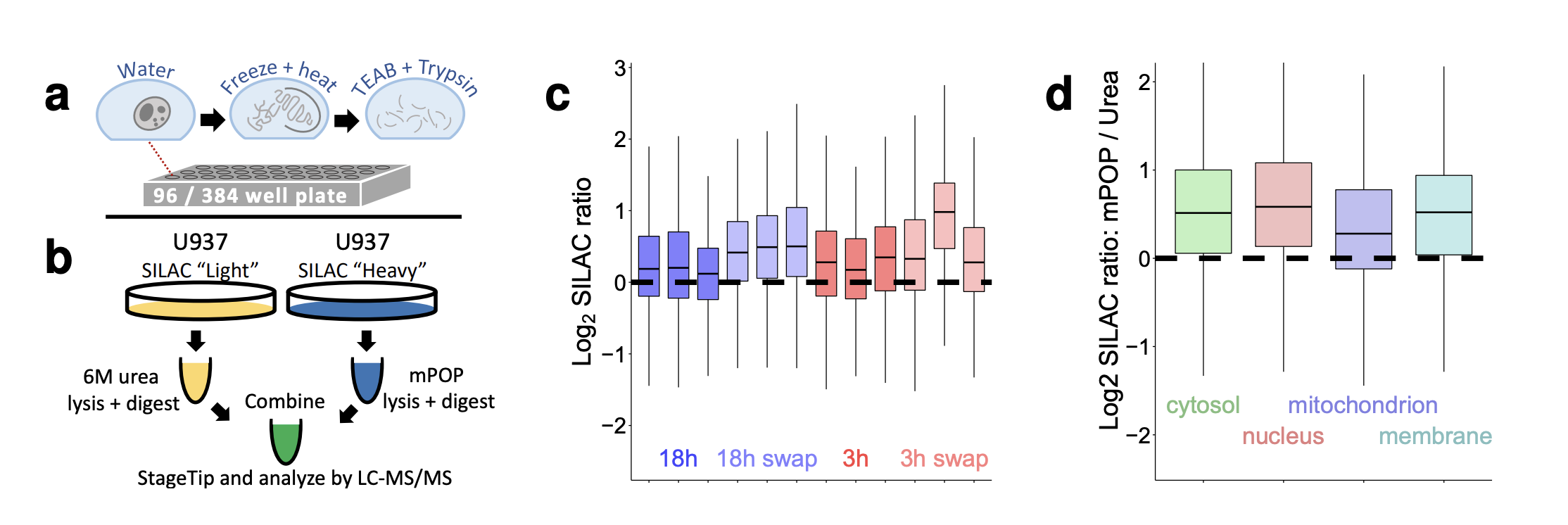
A major limitation to applying quantitative LC-MS/MS proteomics to small samples, such as single cells, are the losses incurred during sample cleanup. To relieve this limitation, Specht et al., 2018 developed Minimal ProteOmic sample Preparation (mPOP). mPOP lyses cells by a freeze-heat cycle and uses only mass-spectrometry compatible chemicals, thus obviating sample cleanup. This approach expediting sample preparation in multi-well plates and simplifies its automation. Bulk SILAC samples processed by mPOP or by conventional urea-based methods demonstrated that mPOP results in complete, unbiased cell lysis and accurate relative quantification. mPOP can be easily intergraded with all methods for single-cell proteomics by mass-spectrometry. Details of its integration with SCoPE2 are described in a Nature protocol and a JoVE video protocol.
Advantages of mPOP
- mPOP uses equipment found in most laboratories
- Easy to adopt and implement without specialized equipment
- mPOP uses only mass-spec compatible chemicals
- No cleanup steps are needed
- mPOP is easy to automate
- Thus mPOP can prepare hundreds of single cells with little human handling
- mPOP uses inexpensive and accessible consumables
- This keeps costs below 1 USD / single cell.
The least reliable step of mPOP is single cell isolation via FACS sorting. The reliability of this step is dependent on the sorters and operators and can be high with experienced operators. Single cells can also be isolated by laser capture microdissection, manual cell picking, CellenONE, magnetic-activated cell sorting, and other methods. We have observed highly consistent and reliable results from mPOP when isolating single cells with CellenONE.
Applications & data
- Specht et al, 2019 used mPOP to prepare samples for SCopE2 analysis of the emergence of macrophage polarization. Specht et al, 2019 data website
- Petelski et al, 2021 demonstrated mPOP as part of a Nature Protocol for SCopE2 analysis. Petelski et al, 2021 data website
- Specht et al, 2018 developed mPOP and below are links to the data from benchmarking experiments demonstrating that:
- mPOP efficiently extracts proteins from all cellular compartments and delivers them for mass-spec analysis
- mPOP supports accurate and precise protein quantification consistent with quantification obtained from standard urea lysis
Recorded video presentations
YouTube Playlist
About the project
This project on developing an automated sample preparation method for single-cell proteomics was supported by funding from the NIH Director’s Award.