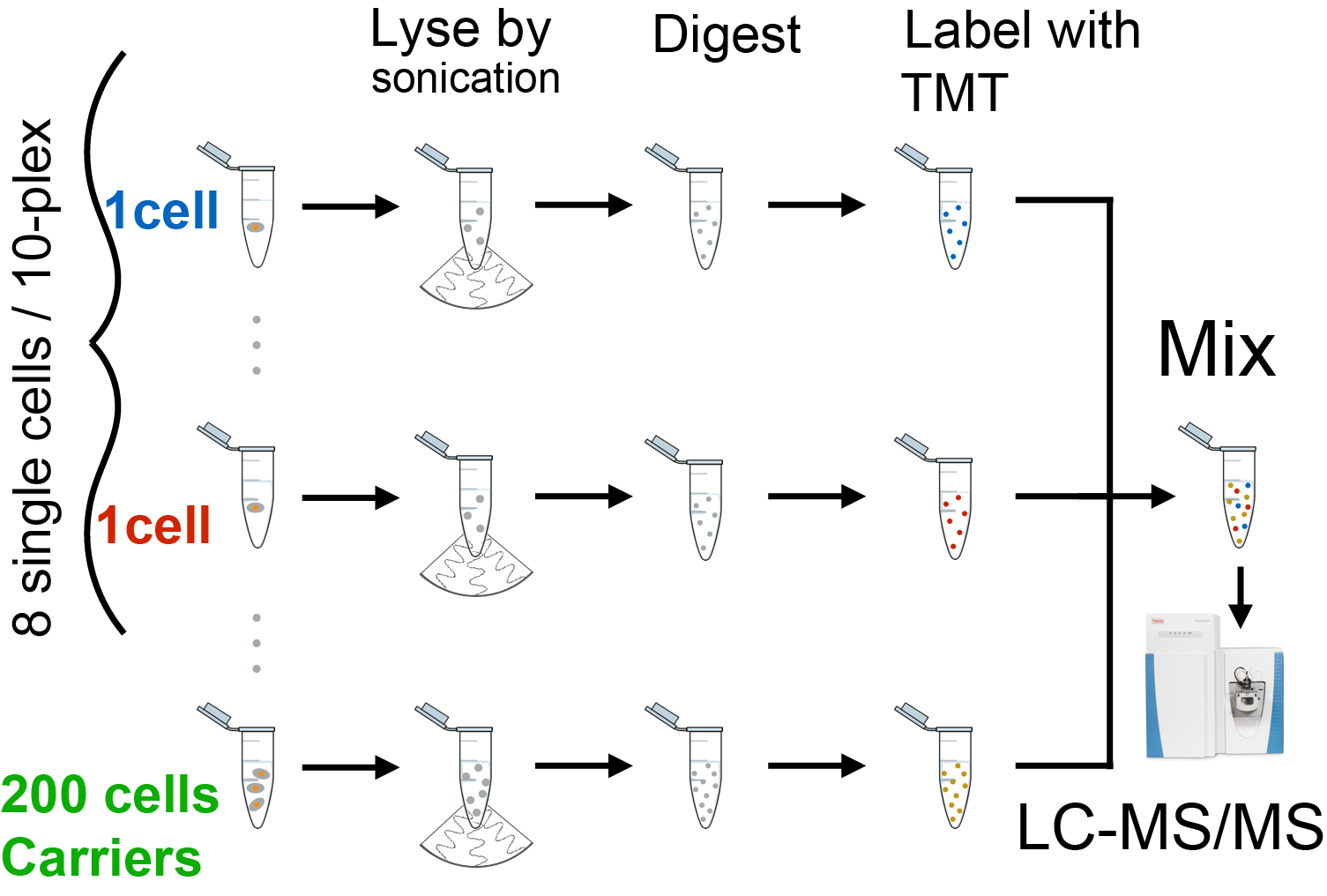
SCoPE-MS
Shotgun single-cell proteomics method by Budnik et al., 2017
Peer reviewed article: Budnik, B., Levy, E., Harmange, G., and Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 19, 161 (2018). 10.1186/s13059-018-1547-5
Data Websites
Some exciting biological questions require quantifying thousands of proteins in single cells. To achieve this goal, we develop Single Cell ProtEomics by Mass Spectrometry (SCoPE-MS) and validate its ability to identify distinct human cancer cell types based on their proteomes. We use SCoPE-MS to quantify over a thousand proteins in differentiating mouse embryonic stem cells. The single-cell proteomes enable us to deconstruct cell populations and infer protein abundance relationships. Comparison between single-cell proteomes and transcriptomes indicates coordinated mRNA and protein covariation, yet many genes exhibit functionally concerted and distinct regulatory patterns at the mRNA and the protein level.