Droplet proteomic sample preparation (nPOP)
Massively parallel sample prep method by Leduc et al, 2021
Peer reviewed article: Leduc A, Huffman RG, Cantlon J, Khan S, Slavov N Exploring functional protein covariation across single cells using nPOP Genome Biol 23, 261 10.1186/s13059-022-02817-5, Data Websites
Protocol article: Leduc A, Koury L, Cantlon J, Slavov N Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP, bioRxiv 2023.11.27.568927
Research Article Protocols GitHub nPOP videos
Data Websites
- Leduc et al., 2021
- Derks et al., 2022
- Leduc et al., 2022
- Huffman et al., 2022
- Montalvo et al., 2023
- Leduc et al., 2023
- Khan et al., 2023
Introduction Resources Advantages of nPOP Applications Videos
Introduction
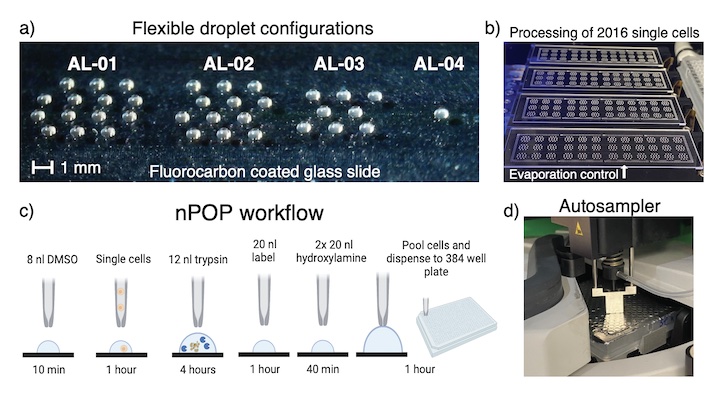
The nano-ProteOmic sample Preparation (nPOP) uses piezo acoustic dispensing to isolate individual cells in 300 picoliter volumes and performs all subsequent preparation steps in small droplets on a fluorocarbon-coated slide. This design enables simultaneous sample preparation of thousands of single cells, including lysing, digesting, and labeling individual cells in volumes below 20 nl.
Advantages of nPOP
- nPOP can prepare thousands of single cells in a single batch
- This minimizes the # of batches and associated technical variation
- nPOP uses only mass-spec compatible chemicals and volumes below 20 nl
- This results in low background and low contamination
- nPOP droplet layouts are computationally programmable and thus very flexible
- Thus nPOP can be used for all multiplexed or label-free workflows without specialized consumables
- nPOP uses accessible consumables and small amounts of labels
- This keeps costs down to about 10-20 cents / single cell.
nPOP uses the CellenONE instrument, which is commercially available. Without access to CellenONE, one may use mPOP, which can prepares a few hundred single cells in parallel. Both mPOP and nPOP are fully compatible with all mass-spectrometry methods described in this portal.
Applications & data
- Leduc et al, 2021 used nPOP to prepare samples for SCopE2 analysis of the cell division cycle of U937 and Hela cells. Leduc et al, 2021 data website
- Derks et al, 2022 used nPOP to prepare samples for plexDIA analysis of single melanoma, monocyte and pancreatic adenocarcinoma cells. Derks et al, 2022 data website
- Leduc et al, 2022 used nPOP to prepare samples for pSCopE analysis of protein covariation in monocytes and melanoma cells. Leduc et al, 2022 data website
- Huffman et al, 2022 used nPOP to prepare samples for pSCopE analysis of primary macrophages stimulated with LPS. Huffman et al, 2022 data website
- Leduc et al, 2023 used nPOP to prepare samples for plexDIA analysis of single melanoma, monocyte and pancreatic adenocarcinoma cells. Leduc et al, 2023 data website
- Khan et al, 2023 used nPOP to prepare samples for SCoPE2 analysis of single epithelial cells undergoing epithelial–mesenchymal transition. Khan et al, 2023 data website
Resources and protocols
-
The current protocol has been preprinted by Leduc et al.. This latest version of the protocol was created in partnership with Cellenion, and the company provides support for implementing nPOP, including required software for the protocol, which can be obtained through our partnership program. Reaching out to Josh Cantlon (j.cantlon@cellenion.com) or Andrew Leduc (leduc.an@northeastern.edu) for more details.
-
The original version of the nPOP protocol can be found here and the cellenONE software files for performing sample prep can be found here.
-
Additionally, tools for mapping single cell image data from the cellenONE to sets run for LC/MS are available here on this GitHub page.
A google group for questions and answers about nPOP.
Recorded video presentations
YouTube Playlist